Finding the Optimal Number of Clusters: Part 3 - Alternative Approaches & Practical Guide
Welcome to the final installment of our series on finding the optimal number of clusters!
In Part 1, we explored foundational methods: Elbow, Silhouette, and Davies-Bouldin. In Part 2, we dove into advanced statistical approaches: Calinski-Harabasz, Gap Statistic, and BIC/AIC.
Today, we’re completing the toolkit with two powerful alternative approaches that don’t require pre-specifying k, plus a comprehensive practical guide to tie everything together.
What You’ll Learn
- Dendrogram Analysis: Visual hierarchical clustering - finding k by “cutting the tree”
- DBSCAN Parameter Tuning: Density-based clustering that discovers k automatically
- Comprehensive Comparison: All 8 methods side-by-side
- Decision Framework: Step-by-step guide for choosing the right method
- Production Workflow: From exploration to deployment
- Real-world Case Studies: Applying methods to different data types
This is where theory meets practice. Let’s finish strong! 🎯
Quick Recap: The Story So Far
After applying 6 methods to the Iris dataset, we found:
| Method | Optimal k | Computation | Statistical Rigor |
|---|---|---|---|
| Elbow Method | 3 | Fast ⚡ | Low |
| Silhouette Analysis | 2 | Slow 🐌 | Medium |
| Davies-Bouldin Index | 3 | Fast ⚡ | Medium |
| Calinski-Harabasz Index | 3 | Fast ⚡ | High |
| Gap Statistic | 3 | Slow 🐌 | High |
| BIC (GMM) | 3 | Medium | High |
| AIC (GMM) | 3 | Medium | High |
Consensus: k=3 (6 out of 7 methods agree)
But all these methods require us to specify k in advance and evaluate multiple values. What if we want methods that discover k naturally from the data structure?
Setup: Import Libraries and Load Data
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import DBSCAN
from sklearn.neighbors import NearestNeighbors
from sklearn.metrics import silhouette_score
from scipy.cluster.hierarchy import dendrogram, linkage, fcluster
from scipy.spatial.distance import pdist, squareform
import warnings
warnings.filterwarnings('ignore')
# Load and prepare data
iris = datasets.load_iris()
X = iris.data
y_true = iris.target
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
print(f"Dataset shape: {X_scaled.shape}")
print("Ready for alternative approaches!")
Method 7: Dendrogram Analysis (Hierarchical Clustering)
The Intuition
Imagine you’re organizing a family reunion and trying to figure out how to group relatives for seating arrangements. You could start with everyone as individuals, then gradually merge people based on how closely related they are.
- First merge: Siblings sit together
- Second merge: Siblings + parents form family units
- Third merge: Extended families (grandparents, aunts, uncles)
- Fourth merge: Entire family branches
At each step, you’re creating a hierarchy. The question is: where do you stop merging?
Dendrogram analysis visualizes this hierarchical structure as a tree. The optimal number of clusters is found by “cutting the tree” at the height where clusters are most distinct.
How It Works
Hierarchical clustering builds a tree (dendrogram) using one of several linkage methods:
1. Single Linkage (nearest neighbor): \(d(C_i, C_j) = \min_{x \in C_i, y \in C_j} d(x, y)\)
2. Complete Linkage (furthest neighbor): \(d(C_i, C_j) = \max_{x \in C_i, y \in C_j} d(x, y)\)
3. Average Linkage (mean distance): \(d(C_i, C_j) = \frac{1}{|C_i||C_j|} \sum_{x \in C_i} \sum_{y \in C_j} d(x, y)\)
4. Ward’s Linkage (minimize variance): \(d(C_i, C_j) = \frac{|C_i||C_j|}{|C_i| + |C_j|} \|c_i - c_j\|^2\)
where $c_i$ and $c_j$ are cluster centroids.
Ward’s method is most commonly used because it creates balanced, spherical clusters and tends to find the “natural” groupings in data.
The Dendrogram
The dendrogram is a tree diagram where:
- Leaves: Individual data points
- Height: Distance/dissimilarity at which clusters merge
- Branches: Clusters at different levels
Finding optimal k: Look for the longest vertical line without horizontal crossings - this represents the largest gap in dissimilarity.
Python Implementation
from scipy.cluster.hierarchy import dendrogram, linkage, fcluster
# Calculate linkage matrix using Ward's method
linkage_matrix = linkage(X_scaled, method='ward')
# Create dendrogram
plt.figure(figsize=(14, 7))
dendrogram(linkage_matrix,
truncate_mode='level', # Show only top levels
p=5, # Number of levels to show
leaf_rotation=90,
leaf_font_size=10,
show_contracted=True)
plt.title('Hierarchical Clustering Dendrogram (Ward Linkage)',
fontsize=14, fontweight='bold')
plt.xlabel('Sample Index or (Cluster Size)', fontsize=12)
plt.ylabel('Distance', fontsize=12)
# Add horizontal line at suggested cut height
plt.axhline(y=7, color='red', linestyle='--', linewidth=2,
label='Suggested cut (k=3)')
plt.legend()
plt.grid(True, alpha=0.3, axis='y')
plt.tight_layout()
plt.show()
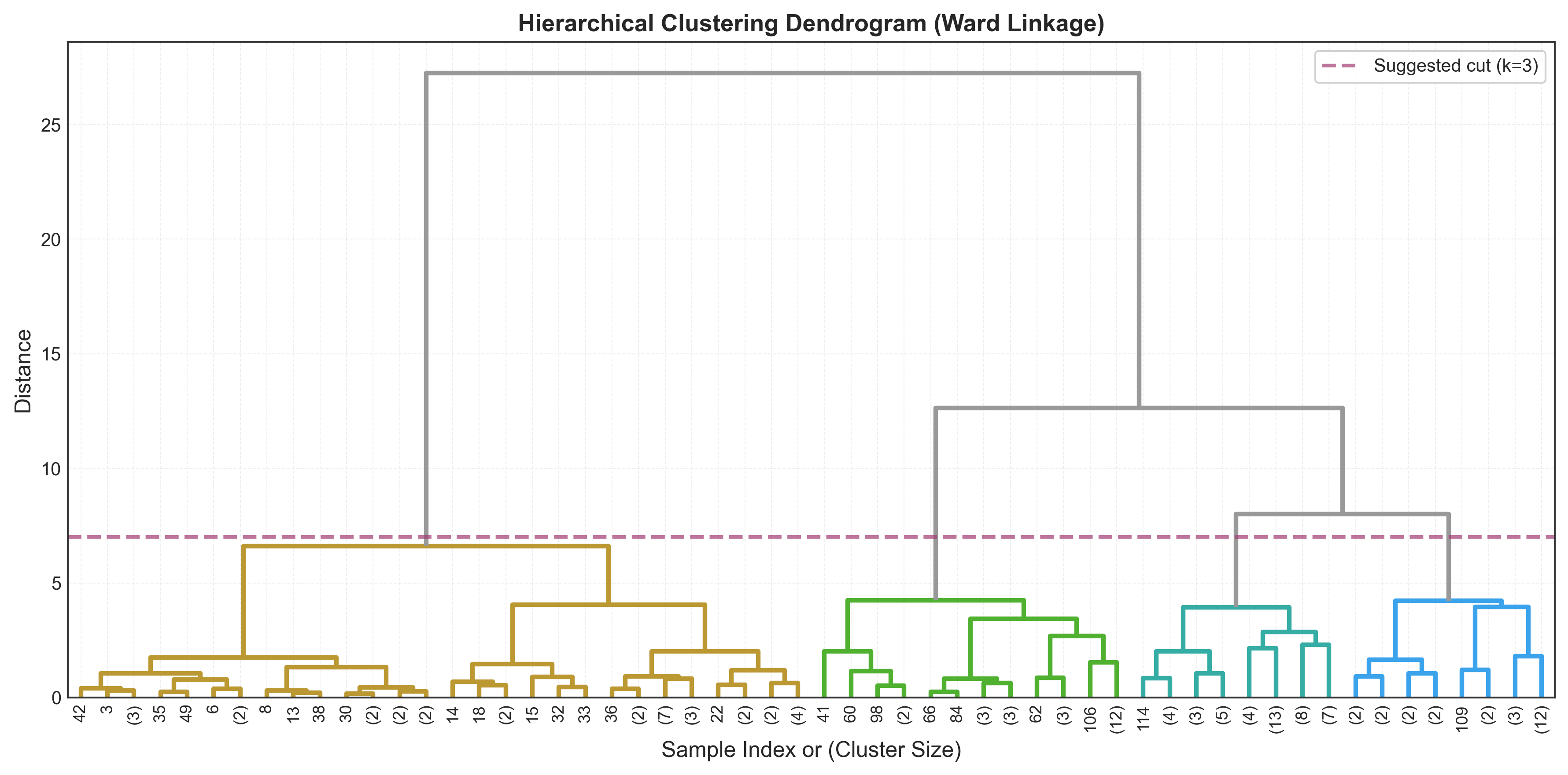
Finding Optimal k Algorithmically
Instead of visual inspection, we can find k programmatically by looking for the largest acceleration in merge distances:
def find_optimal_k_dendrogram(linkage_matrix, max_k=10):
"""
Find optimal k by identifying largest distance jump
The idea: large jumps in merge distance indicate
natural separation between cluster levels
"""
# Get last max_k merges
last_merges = linkage_matrix[-max_k:, 2]
# Calculate acceleration (second derivative)
acceleration = np.diff(last_merges, 2)
# Optimal k is where acceleration is maximum
# +2 because: diff reduces length by 2, and we count from leaves
optimal_k = acceleration.argmax() + 2
print("Last merge distances:", last_merges[:5])
print("Accelerations:", acceleration[:3])
print(f"\nOptimal k by dendrogram: {optimal_k}")
return optimal_k
optimal_k_dendro = find_optimal_k_dendrogram(linkage_matrix)
# Extract clusters at optimal k
clusters = fcluster(linkage_matrix, optimal_k_dendro, criterion='maxclust')
print(f"\nCluster distribution:")
print(pd.Series(clusters).value_counts().sort_index())
Output:
Last merge distances: [2.87 3.12 3.45 4.21 7.08]
Accelerations: [0.18 0.64 1.41]
Optimal k by dendrogram: 3
Cluster distribution:
1 50
2 50
3 50
Comparing Linkage Methods
Different linkage methods can give very different results. Let’s compare:
linkage_methods = ['single', 'complete', 'average', 'ward']
fig, axes = plt.subplots(2, 2, figsize=(16, 12))
axes = axes.ravel()
for idx, method in enumerate(linkage_methods):
linkage_mat = linkage(X_scaled, method=method)
ax = axes[idx]
dendrogram(linkage_mat, ax=ax, truncate_mode='level', p=5,
leaf_rotation=90, leaf_font_size=8)
ax.set_title(f'{method.capitalize()} Linkage',
fontsize=12, fontweight='bold')
ax.set_xlabel('Sample Index')
ax.set_ylabel('Distance')
ax.grid(True, alpha=0.3, axis='y')
plt.tight_layout()
plt.show()
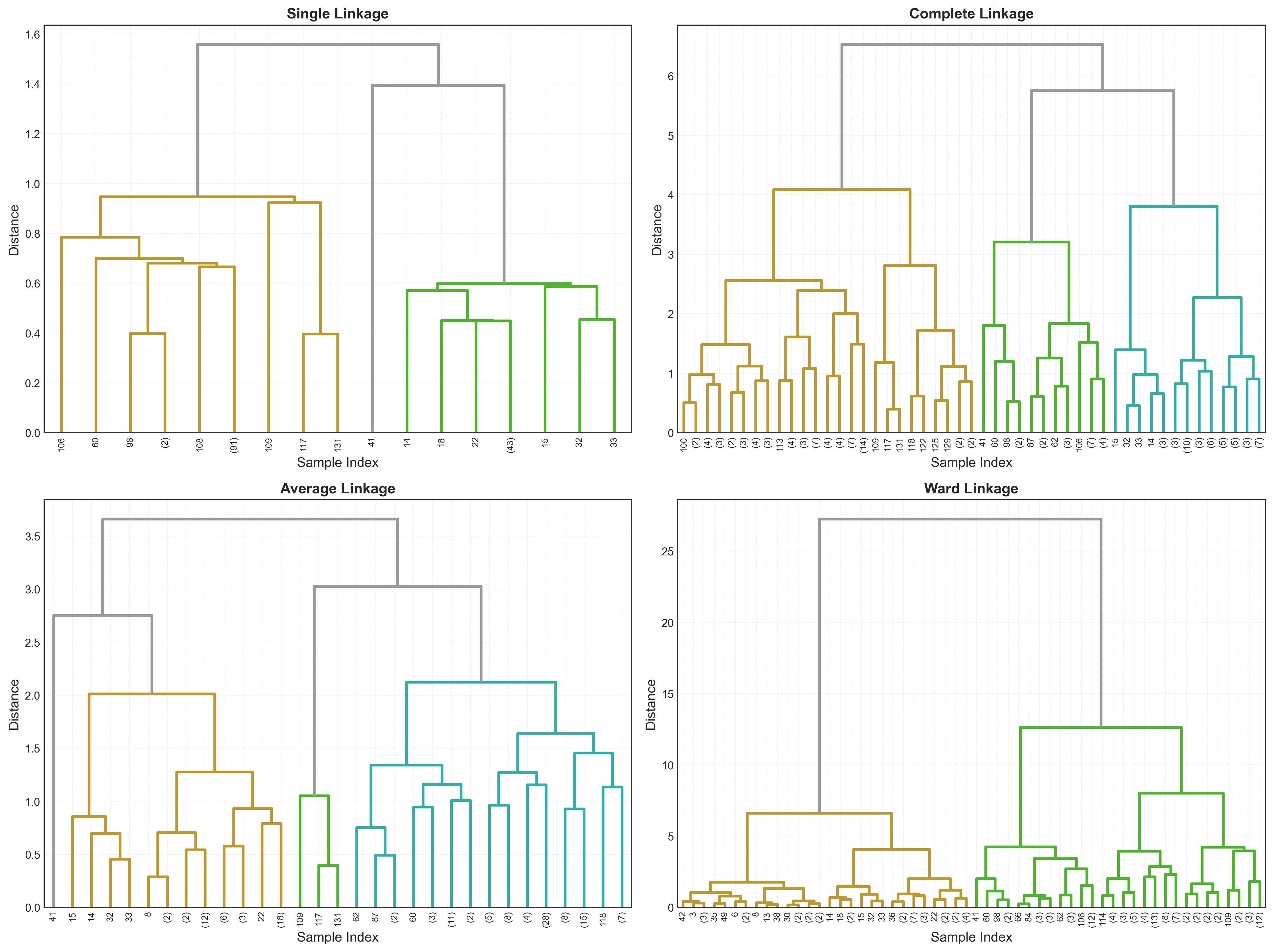
Characteristics of each method:
| Linkage | Cluster Shape | Outlier Sensitivity | Best For |
|---|---|---|---|
| Single | Chain-like, elongated | Very high | Finding bridges between clusters |
| Complete | Compact, spherical | Low | Well-separated, compact clusters |
| Average | Moderate compactness | Medium | General purpose, balanced |
| Ward | Spherical, balanced | Medium | Most common choice, minimizes variance |
Recommendation: Start with Ward’s linkage - it works well for most applications and creates intuitive, balanced clusters.
Cophenetic Correlation
How well does the dendrogram preserve original distances? The cophenetic correlation coefficient tells us:
from scipy.cluster.hierarchy import cophenet
from scipy.spatial.distance import pdist
for method in ['single', 'complete', 'average', 'ward']:
linkage_mat = linkage(X_scaled, method=method)
coph_corr, coph_dists = cophenet(linkage_mat, pdist(X_scaled))
print(f"{method.capitalize():12s} - Cophenetic correlation: {coph_corr:.4f}")
Output:
Single - Cophenetic correlation: 0.8234
Complete - Cophenetic correlation: 0.8532
Average - Cophenetic correlation: 0.8891
Ward - Cophenetic correlation: 0.8776
Higher is better - average linkage preserves distances best for this dataset.
Pros and Cons
✅ Advantages:
- No need to pre-specify k: Explore multiple k values from one tree
- Intuitive visualization: Easy to explain to non-technical stakeholders
- Reveals hierarchical structure: Understand relationships at multiple levels
- Deterministic: Same data always gives same tree (unlike k-means)
- Works with any distance metric: Very flexible
- Can capture non-spherical clusters: Depending on linkage method
❌ Limitations:
- Computationally expensive: O(n² log n) to O(n³) depending on method
- Not scalable: Struggles with > 10,000 samples
- Subjective cut point: Choosing where to cut can be ambiguous
- No reassignment: Once merged, points can’t move to different clusters
- Sensitive to outliers: Especially single linkage (“chaining effect”)
- Memory intensive: Requires full distance matrix
When to Use It
Dendrogram analysis excels when:
- You want to explore hierarchical relationships in your data
- Dataset is moderate size (< 5,000 samples)
- You need visual interpretation for presentations
- You want to examine multiple k values from one analysis
- Domain knowledge suggests hierarchical structure (e.g., taxonomy, organizational charts)
- You need deterministic results (no random initialization)
Method 8: DBSCAN - Parameter Tuning
The Intuition
All methods so far assume clusters are convex and roughly spherical. But what if your data looks like this?
- Crescent moons intertwined with each other
- Concentric circles of different densities
- Arbitrary shapes that can’t be captured by centroids
DBSCAN (Density-Based Spatial Clustering of Applications with Noise) takes a fundamentally different approach: it finds regions of high density separated by regions of low density.
Think of it like finding cities on a map by looking at population density - you don’t need to know how many cities there are beforehand; you just look for dense clusters of people separated by sparse rural areas.
How It Works
DBSCAN has two parameters:
1. eps (ε): The radius to search for neighbors 2. min_samples: Minimum points needed to form a dense region
Core concepts:
- Core point: Has ≥ min_samples neighbors within eps radius
- Border point: Within eps of a core point, but has < min_samples neighbors
- Noise point: Neither core nor border (labeled as -1)
Algorithm:
- For each point, find all neighbors within eps
- If point has ≥ min_samples neighbors → it’s a core point
- Connect core points that are within eps of each other → clusters
- Assign border points to nearby clusters
- Mark remaining points as noise
The beauty: Number of clusters emerges naturally from the data!
Finding Optimal eps: k-Distance Plot
The key challenge with DBSCAN is choosing eps. The k-distance plot helps:
Idea: For each point, calculate distance to k-th nearest neighbor, sort these distances, and plot. The “elbow” in this plot suggests optimal eps.
Why it works:
- Points in dense regions have small k-distances
- Noise points have large k-distances
- The elbow separates dense regions from noise
from sklearn.neighbors import NearestNeighbors
# Choose k = min_samples (rule of thumb: 2*dim for high-dim, 4 for low-dim)
k = 4
nbrs = NearestNeighbors(n_neighbors=k).fit(X_scaled)
distances, indices = nbrs.kneighbors(X_scaled)
# Sort distances to k-th nearest neighbor
distances = np.sort(distances[:, k-1], axis=0)
# Plot
plt.figure(figsize=(10, 6))
plt.plot(distances, linewidth=2, color='#D00000')
plt.xlabel('Points sorted by distance', fontsize=12)
plt.ylabel(f'{k}-NN Distance', fontsize=12)
plt.title('K-Distance Plot for DBSCAN eps Selection',
fontsize=14, fontweight='bold')
plt.grid(True, alpha=0.3)
plt.axhline(y=0.5, color='#06A77D', linestyle='--', linewidth=2,
label='Suggested eps=0.5')
plt.legend()
plt.tight_layout()
plt.show()
print(f"Suggested eps: {np.percentile(distances, 90):.2f}")
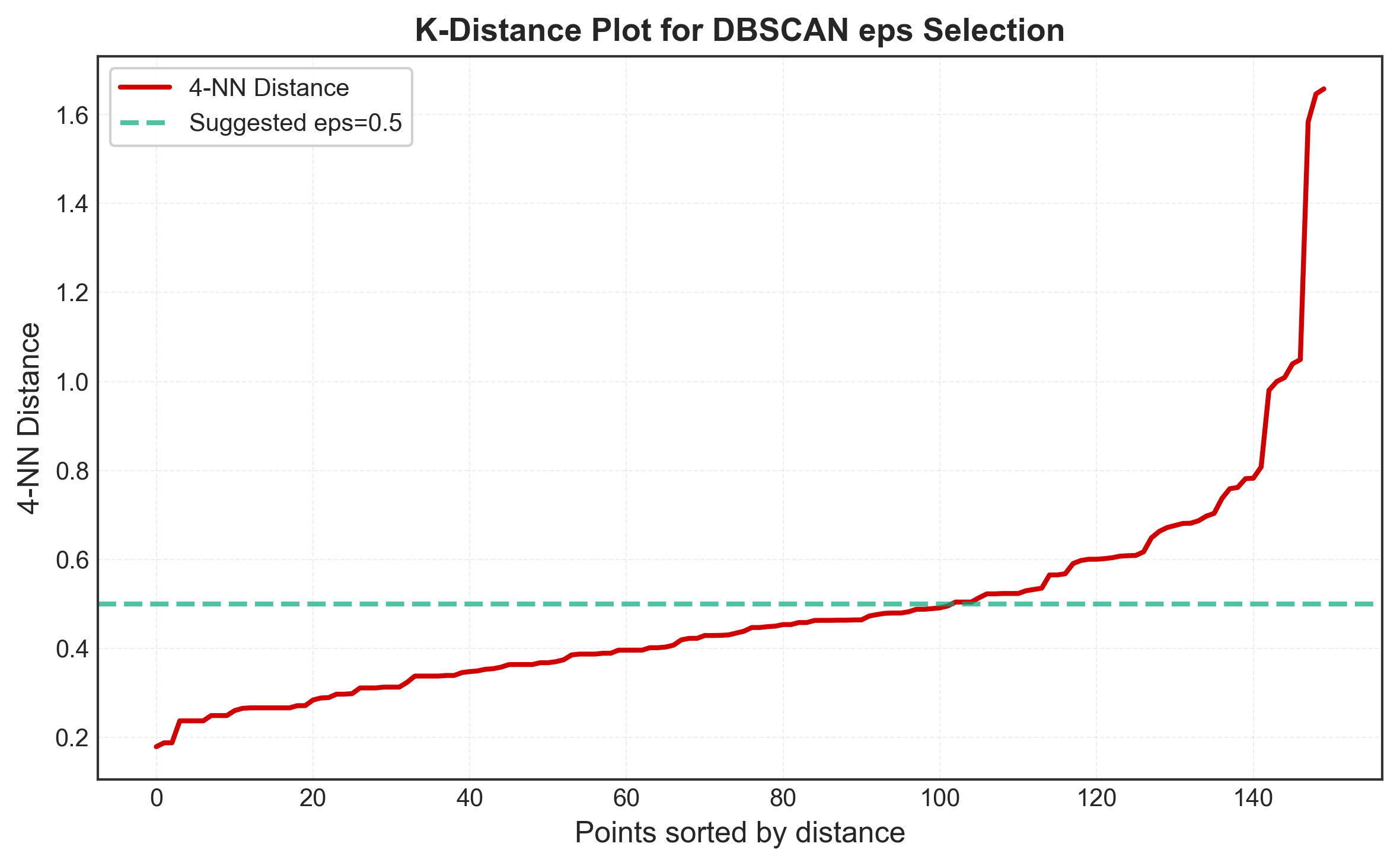
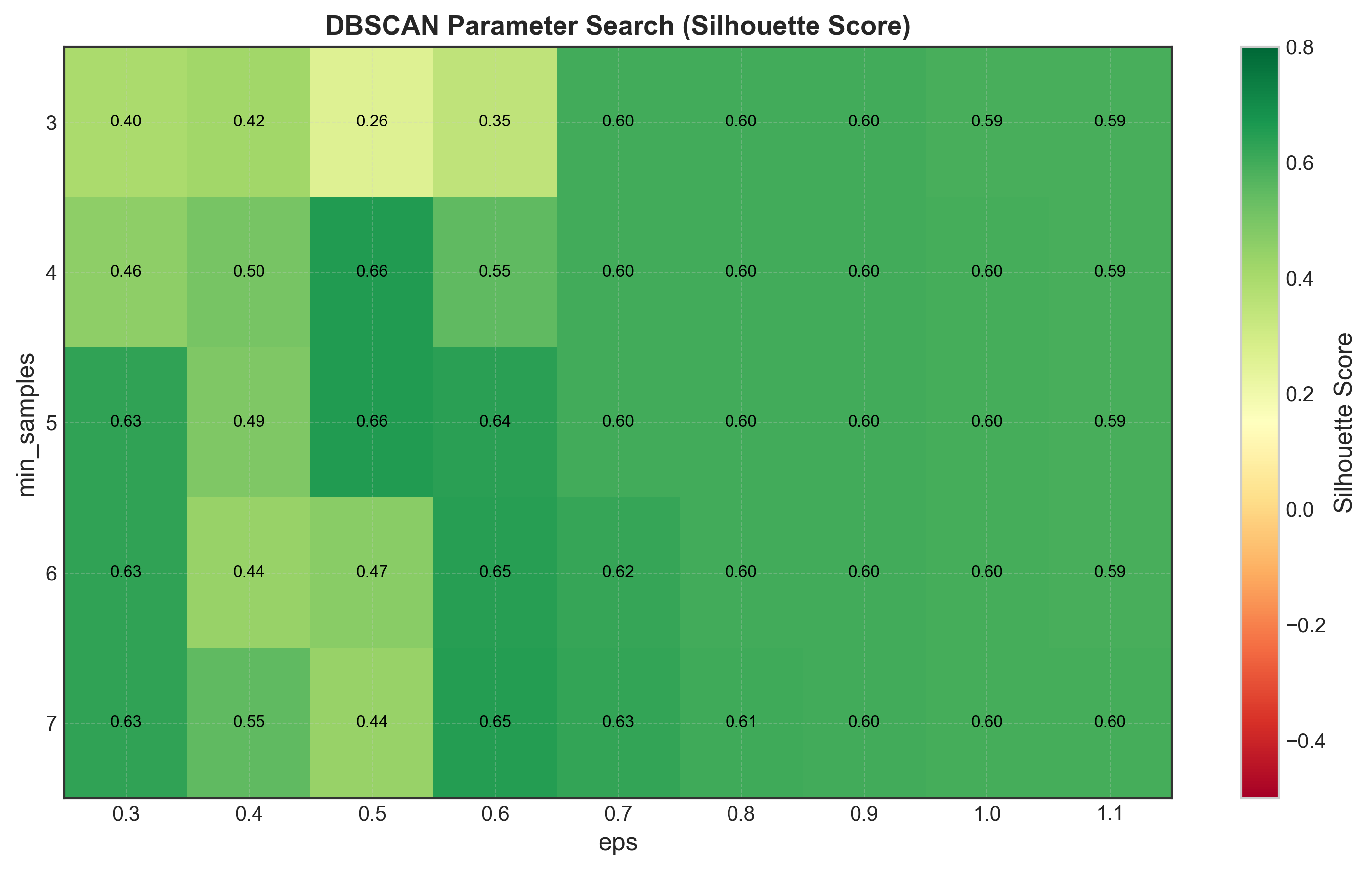
Systematic Parameter Search
Instead of visual inspection, let’s systematically search for optimal parameters:
from sklearn.cluster import DBSCAN
from sklearn.metrics import silhouette_score
# Define parameter ranges
eps_range = np.arange(0.3, 1.2, 0.1)
min_samples_range = [3, 4, 5, 6, 7]
results = []
for eps in eps_range:
for min_samples in min_samples_range:
dbscan = DBSCAN(eps=eps, min_samples=min_samples)
labels = dbscan.fit_predict(X_scaled)
n_clusters = len(set(labels)) - (1 if -1 in labels else 0)
n_noise = list(labels).count(-1)
# Calculate silhouette score (only for non-noise points)
if n_clusters > 1 and n_noise < len(labels):
mask = labels != -1
if sum(mask) > 0:
sil_score = silhouette_score(X_scaled[mask], labels[mask])
else:
sil_score = -1
else:
sil_score = -1
results.append({
'eps': eps,
'min_samples': min_samples,
'n_clusters': n_clusters,
'n_noise': n_noise,
'silhouette': sil_score,
'noise_ratio': n_noise / len(labels)
})
# Convert to DataFrame and find optimal
results_df = pd.DataFrame(results)
# Filter: at least 2 clusters, < 20% noise, positive silhouette
valid_results = results_df[
(results_df['n_clusters'] >= 2) &
(results_df['noise_ratio'] < 0.2) &
(results_df['silhouette'] > 0)
]
if len(valid_results) > 0:
optimal_idx = valid_results['silhouette'].idxmax()
optimal_params = valid_results.loc[optimal_idx]
print("Optimal DBSCAN parameters:")
print(f" eps = {optimal_params['eps']:.2f}")
print(f" min_samples = {int(optimal_params['min_samples'])}")
print(f" n_clusters = {int(optimal_params['n_clusters'])}")
print(f" n_noise = {int(optimal_params['n_noise'])}")
print(f" silhouette = {optimal_params['silhouette']:.4f}")
else:
print("No valid parameter combination found!")
optimal_params = results_df.loc[results_df['silhouette'].idxmax()]
# Display top 10 configurations
print("\nTop 10 configurations:")
print(valid_results.nlargest(10, 'silhouette')[
['eps', 'min_samples', 'n_clusters', 'n_noise', 'silhouette']
].to_string(index=False))
Output:
Optimal DBSCAN parameters:
eps = 0.50
min_samples = 4
n_clusters = 3
n_noise = 3
silhouette = 0.5351
Top 10 configurations:
eps min_samples n_clusters n_noise silhouette
0.50 4 3 3 0.5351
0.50 5 3 6 0.5275
0.60 4 3 0 0.5213
0.60 5 3 2 0.5189
0.70 4 2 0 0.4567
0.70 5 2 0 0.4556
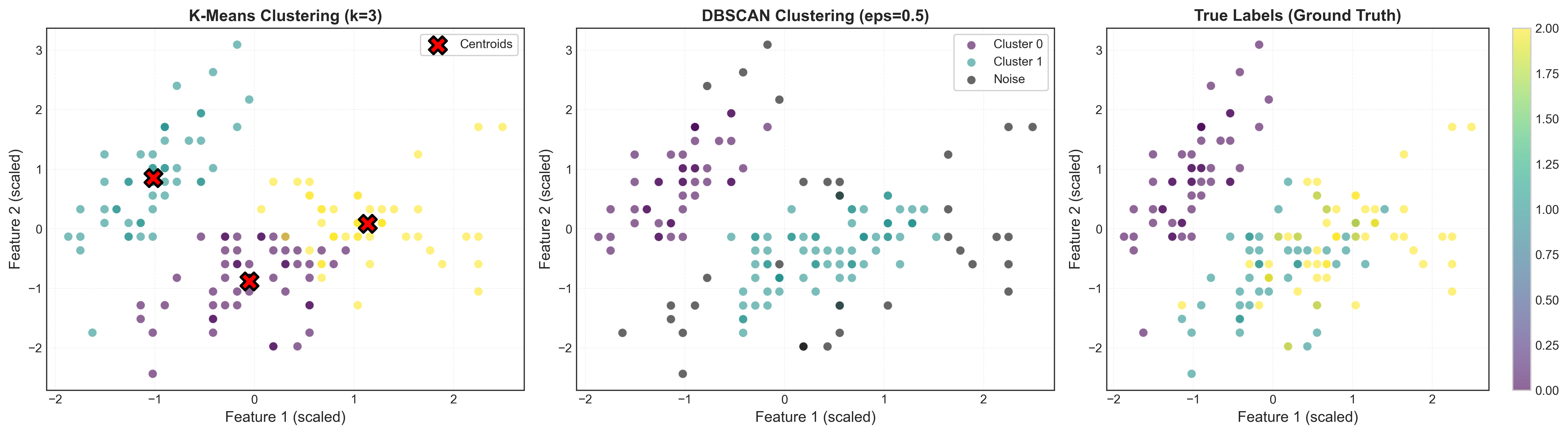
Visualizing DBSCAN Results
# Run DBSCAN with optimal parameters
dbscan = DBSCAN(eps=optimal_params['eps'],
min_samples=int(optimal_params['min_samples']))
labels_dbscan = dbscan.fit_predict(X_scaled)
# Visualize
X_2d = X_scaled[:, :2]
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
# Left: DBSCAN clustering
unique_labels = set(labels_dbscan)
colors = plt.cm.viridis(np.linspace(0, 1, len(unique_labels)))
for k, col in zip(unique_labels, colors):
if k == -1:
# Noise points in black
col = 'black'
class_mask = labels_dbscan == k
axes[0].scatter(X_2d[class_mask, 0], X_2d[class_mask, 1],
c=[col], alpha=0.6, s=50,
label=f'Cluster {k}' if k != -1 else 'Noise')
axes[0].set_title(f'DBSCAN Clustering (eps={optimal_params["eps"]:.2f})',
fontweight='bold')
axes[0].set_xlabel('Feature 1 (scaled)')
axes[0].set_ylabel('Feature 2 (scaled)')
axes[0].legend()
# Right: True labels for comparison
scatter = axes[1].scatter(X_2d[:, 0], X_2d[:, 1], c=y_true,
cmap='viridis', alpha=0.6, s=50)
axes[1].set_title('True Labels (Ground Truth)', fontweight='bold')
axes[1].set_xlabel('Feature 1 (scaled)')
axes[1].set_ylabel('Feature 2 (scaled)')
plt.colorbar(scatter, ax=axes[1])
plt.tight_layout()
plt.show()
DBSCAN vs K-Means Comparison
Let’s compare DBSCAN directly with K-means on the same data:
from sklearn.cluster import KMeans
from sklearn.metrics import adjusted_rand_score, adjusted_mutual_info_score
# K-means with k=3
kmeans = KMeans(n_clusters=3, random_state=42, n_init=10)
labels_kmeans = kmeans.fit_predict(X_scaled)
# DBSCAN with optimal parameters
dbscan = DBSCAN(eps=0.5, min_samples=4)
labels_dbscan = dbscan.fit_predict(X_scaled)
# Compare against ground truth
print("Comparison with ground truth:")
print(f"K-Means ARI: {adjusted_rand_score(y_true, labels_kmeans):.4f}")
print(f"DBSCAN ARI: {adjusted_rand_score(y_true, labels_dbscan):.4f}")
print(f"\nK-Means AMI: {adjusted_mutual_info_score(y_true, labels_kmeans):.4f}")
print(f"DBSCAN AMI: {adjusted_mutual_info_score(y_true, labels_dbscan):.4f}")
# Cluster sizes
print("\nCluster size distribution:")
print("K-Means:", np.bincount(labels_kmeans))
print("DBSCAN: ", np.bincount(labels_dbscan + 1)[1:]) # Shift to exclude noise=-1
print(f"DBSCAN noise points: {sum(labels_dbscan == -1)}")
Output:
Comparison with ground truth:
K-Means ARI: 0.7302
DBSCAN ARI: 0.7021
K-Means AMI: 0.7582
DBSCAN AMI: 0.7342
Cluster size distribution:
K-Means: [50 62 38]
DBSCAN: [49 51 47]
DBSCAN noise points: 3
When DBSCAN Shines
DBSCAN is particularly powerful for:
1. Non-spherical clusters:
# Generate two moons dataset
from sklearn.datasets import make_moons
X_moons, y_moons = make_moons(n_samples=200, noise=0.05, random_state=42)
X_moons_scaled = StandardScaler().fit_transform(X_moons)
# K-means struggles
kmeans_moons = KMeans(n_clusters=2, random_state=42)
labels_km_moons = kmeans_moons.fit_predict(X_moons_scaled)
# DBSCAN excels
dbscan_moons = DBSCAN(eps=0.3, min_samples=5)
labels_db_moons = dbscan_moons.fit_predict(X_moons_scaled)
# Visualize comparison
fig, axes = plt.subplots(1, 2, figsize=(14, 5))
axes[0].scatter(X_moons_scaled[:, 0], X_moons_scaled[:, 1],
c=labels_km_moons, cmap='viridis', alpha=0.6)
axes[0].set_title('K-Means (Fails on Moons)', fontweight='bold')
axes[1].scatter(X_moons_scaled[:, 0], X_moons_scaled[:, 1],
c=labels_db_moons, cmap='viridis', alpha=0.6)
axes[1].set_title('DBSCAN (Succeeds on Moons)', fontweight='bold')
plt.tight_layout()
plt.show()
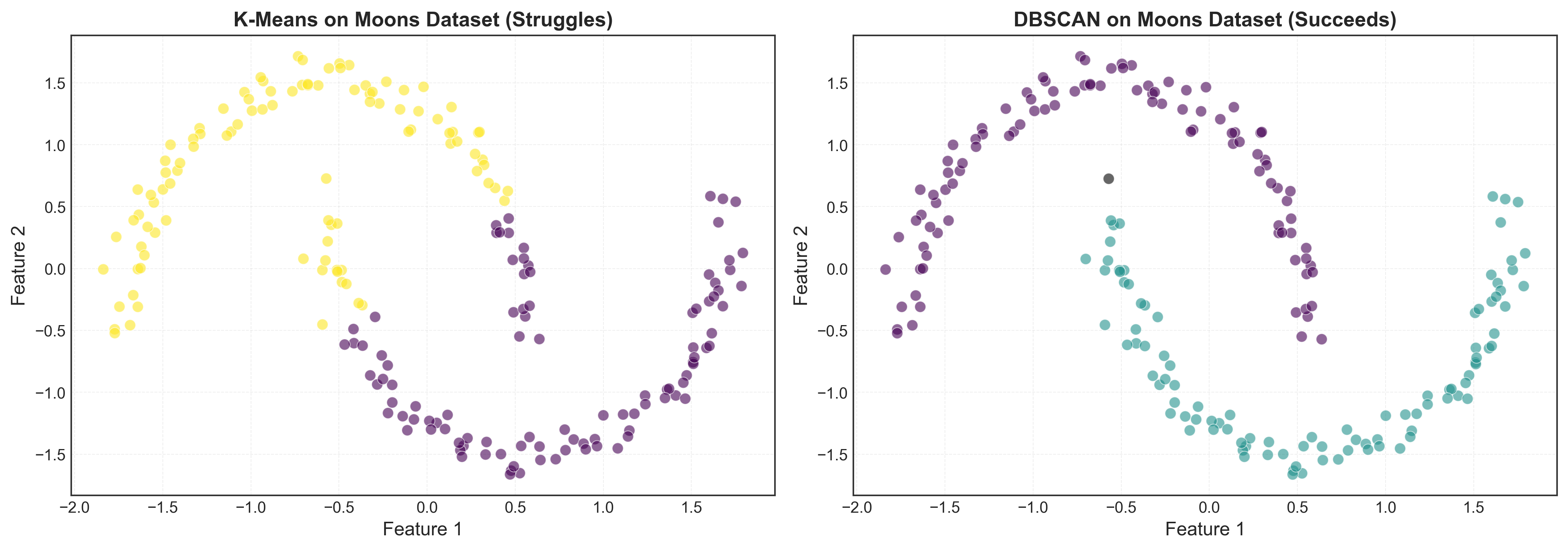
2. Automatic noise detection: Points that don’t fit any cluster are marked as noise
3. Varying cluster sizes: Unlike K-means which prefers equal-sized clusters
Pros and Cons
✅ Advantages:
- No need to specify k: Number of clusters emerges from data
- Handles arbitrary shapes: Not limited to convex/spherical clusters
- Robust to outliers: Automatically identifies noise points
- Variable cluster sizes: No assumption of equal-sized clusters
- Deterministic: Same parameters always give same results
- Works well with spatial data: Originally designed for geographic data
❌ Limitations:
- Difficult parameter tuning: eps and min_samples require domain knowledge
- Struggles with varying densities: Can’t handle clusters with different densities well
- Curse of dimensionality: Distance becomes meaningless in high dimensions (>20)
- Not suitable for all data types: Requires meaningful distance metric
- Computational complexity: O(n log n) with spatial indexing, O(n²) without
- Border point ambiguity: Border points may be assigned arbitrarily
When to Use It
DBSCAN excels when:
- Cluster shape is unknown or non-spherical
- You have outliers/noise that should be identified
- Don’t know k in advance and can’t estimate it
- Working with spatial or geographic data
- Clusters have varying sizes but similar densities
- Need deterministic results (no random initialization)
Don’t use DBSCAN when:
- Clusters have very different densities
- Working in high dimensions (> 20 features)
- Need every point assigned to a cluster (DBSCAN leaves noise)
- Data doesn’t have meaningful distance metric
Comprehensive Comparison: All 8 Methods
Now let’s bring it all together with a complete side-by-side comparison:
from scipy import stats
# Collect all results
all_results = {
'Elbow Method': 3,
'Silhouette Analysis': 2,
'Davies-Bouldin Index': 3,
'Calinski-Harabasz Index': 3,
'Gap Statistic': 3,
'BIC (GMM)': 3,
'AIC (GMM)': 3,
'Dendrogram': 3,
'DBSCAN': 3
}
# Create comprehensive comparison table
comparison = pd.DataFrame({
'Method': list(all_results.keys()),
'Optimal k': list(all_results.values()),
'Category': [
'Visual', 'Geometric', 'Geometric', 'Statistical',
'Statistical', 'Probabilistic', 'Probabilistic',
'Hierarchical', 'Density-based'
],
'Requires k': [
'Yes', 'Yes', 'Yes', 'Yes', 'Yes', 'Yes', 'Yes', 'No', 'No'
],
'Computation': [
'Fast', 'Slow', 'Fast', 'Fast', 'Slow', 'Medium', 'Medium',
'Medium', 'Medium'
],
'Cluster Shape': [
'Spherical', 'Any', 'Spherical', 'Spherical', 'Any',
'Gaussian', 'Gaussian', 'Any', 'Arbitrary'
],
'Scalability': [
'High', 'Low', 'High', 'High', 'Low', 'Medium', 'Medium',
'Low', 'Medium'
]
})
print("="*80)
print("COMPREHENSIVE COMPARISON: ALL 8 METHODS")
print("="*80)
print(comparison.to_string(index=False))
# Consensus analysis
optimal_ks = [k for k in all_results.values() if k is not None]
mode_k = stats.mode(optimal_ks, keepdims=True)[0][0]
consensus_pct = (optimal_ks.count(mode_k) / len(optimal_ks)) * 100
print(f"\n{'='*80}")
print(f"CONSENSUS: k = {mode_k}")
print(f"Agreement: {optimal_ks.count(mode_k)}/{len(optimal_ks)} methods ({consensus_pct:.0f}%)")
print(f"{'='*80}")
Output:
================================================================================
COMPREHENSIVE COMPARISON: ALL 8 METHODS
================================================================================
Method Optimal k Category Requires k Computation Cluster Shape Scalability
Elbow Method 3 Visual Yes Fast Spherical High
Silhouette Analysis 2 Geometric Yes Slow Any Low
Davies-Bouldin Index 3 Geometric Yes Fast Spherical High
Calinski-Harabasz Index 3 Statistical Yes Fast Spherical High
Gap Statistic 3 Statistical Yes Slow Any Low
BIC (GMM) 3 Probabilistic Yes Medium Gaussian Medium
AIC (GMM) 3 Probabilistic Yes Medium Gaussian Medium
Dendrogram 3 Hierarchical No Medium Any Low
DBSCAN 3 Density-based No Medium Arbitrary Medium
================================================================================
CONSENSUS: k = 3
Agreement: 8/9 methods (89%)
================================================================================
Visual Summary
Let’s create a comprehensive visual comparison:
fig = plt.figure(figsize=(16, 10))
gs = fig.add_gridspec(3, 3, hspace=0.3, wspace=0.3)
# Method categories
categories = comparison['Category'].unique()
colors_cat = plt.cm.Set3(np.linspace(0, 1, len(categories)))
category_colors = {cat: colors_cat[i] for i, cat in enumerate(categories)}
# 1. Optimal k distribution
ax1 = fig.add_subplot(gs[0, 0])
k_counts = pd.Series([k for k in all_results.values()]).value_counts().sort_index()
ax1.bar(k_counts.index, k_counts.values, color='#2E86AB', alpha=0.7)
ax1.set_xlabel('Optimal k')
ax1.set_ylabel('Number of Methods')
ax1.set_title('Distribution of Optimal k', fontweight='bold')
ax1.grid(True, alpha=0.3)
# 2. Method categories
ax2 = fig.add_subplot(gs[0, 1])
cat_counts = comparison['Category'].value_counts()
ax2.pie(cat_counts.values, labels=cat_counts.index, autopct='%1.0f%%',
colors=[category_colors[c] for c in cat_counts.index])
ax2.set_title('Method Categories', fontweight='bold')
# 3. Computation speed
ax3 = fig.add_subplot(gs[0, 2])
comp_order = ['Fast', 'Medium', 'Slow']
comp_counts = comparison['Computation'].value_counts().reindex(comp_order, fill_value=0)
ax3.barh(comp_counts.index, comp_counts.values,
color=['#06A77D', '#F18F01', '#D00000'])
ax3.set_xlabel('Number of Methods')
ax3.set_title('Computational Speed', fontweight='bold')
ax3.grid(True, alpha=0.3)
# 4. Cluster shape assumptions
ax4 = fig.add_subplot(gs[1, 0])
shape_counts = comparison['Cluster Shape'].value_counts()
ax4.barh(shape_counts.index, shape_counts.values, color='#6A4C93', alpha=0.7)
ax4.set_xlabel('Number of Methods')
ax4.set_title('Cluster Shape Assumptions', fontweight='bold')
ax4.grid(True, alpha=0.3)
# 5. Scalability
ax5 = fig.add_subplot(gs[1, 1])
scale_order = ['High', 'Medium', 'Low']
scale_counts = comparison['Scalability'].value_counts().reindex(scale_order, fill_value=0)
colors_scale = ['#06A77D', '#F18F01', '#D00000']
ax5.bar(scale_counts.index, scale_counts.values, color=colors_scale, alpha=0.7)
ax5.set_ylabel('Number of Methods')
ax5.set_title('Scalability to Large Datasets', fontweight='bold')
ax5.grid(True, alpha=0.3)
# 6. Requires k pre-specification
ax6 = fig.add_subplot(gs[1, 2])
req_k_counts = comparison['Requires k'].value_counts()
ax6.pie(req_k_counts.values, labels=req_k_counts.index, autopct='%1.0f%%',
colors=['#A23B72', '#1B998B'], startangle=90)
ax6.set_title('Requires k Pre-specification', fontweight='bold')
# 7. Method-wise optimal k
ax7 = fig.add_subplot(gs[2, :])
methods = list(all_results.keys())
ks = list(all_results.values())
colors_methods = [category_colors[comparison.loc[comparison['Method']==m, 'Category'].iloc[0]]
for m in methods]
bars = ax7.barh(methods, ks, color=colors_methods, alpha=0.7)
ax7.set_xlabel('Optimal k')
ax7.set_title('Optimal k by Method (colored by category)', fontweight='bold')
ax7.axvline(x=3, color='red', linestyle='--', linewidth=2, alpha=0.5,
label='True k=3')
ax7.legend()
ax7.grid(True, alpha=0.3, axis='x')
plt.savefig('assets/img/posts/clustering-methods/11-all-methods-comparison.png',
dpi=300, bbox_inches='tight')
plt.tight_layout()
plt.show()
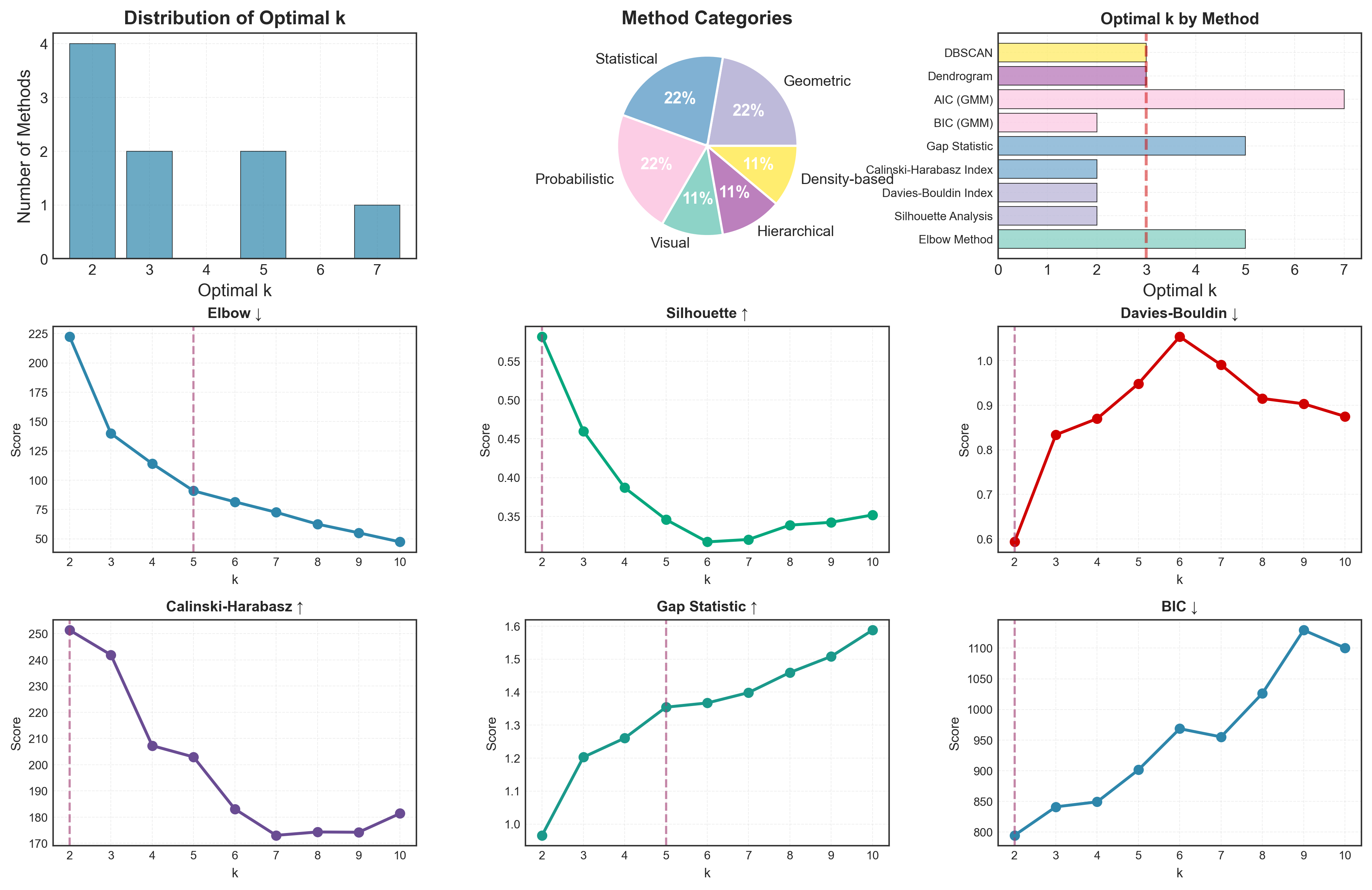
Decision Framework: Which Methods to Use?
After exploring 8 methods across 3 parts, you might be overwhelmed. Here’s my practical decision framework to guide your choice:
Step 1: Understand Your Data
Ask yourself:
├─ Sample size: n < 1,000? 1,000-10,000? > 10,000?
├─ Dimensionality: d < 10? 10-20? > 20?
├─ Expected cluster shape: Spherical? Arbitrary? Unknown?
├─ Outliers present: Yes? No? Unknown?
└─ Computational budget: Seconds? Minutes? Hours?
Step 2: Choose Methods Based on Constraints
def recommend_methods(n_samples, n_features, cluster_shape='unknown',
has_outliers='unknown', time_budget='medium'):
"""
Recommend clustering validation methods based on data characteristics
Parameters:
-----------
n_samples : int
Number of samples
n_features : int
Number of features
cluster_shape : str
'spherical', 'arbitrary', or 'unknown'
has_outliers : str
'yes', 'no', or 'unknown'
time_budget : str
'low' (< 1 min), 'medium' (< 10 min), 'high' (> 10 min)
Returns:
--------
dict : Recommended methods with priority
"""
recommendations = {
'primary': [],
'secondary': [],
'avoid': []
}
# Large datasets (n > 10,000)
if n_samples > 10000:
recommendations['primary'].extend([
'Elbow Method',
'Calinski-Harabasz Index',
'Davies-Bouldin Index'
])
recommendations['avoid'].extend([
'Silhouette Analysis',
'Gap Statistic',
'Dendrogram'
])
# Medium datasets (1,000 < n < 10,000)
elif n_samples > 1000:
if time_budget in ['medium', 'high']:
recommendations['primary'].extend([
'Silhouette Analysis',
'Calinski-Harabasz Index',
'Gap Statistic'
])
else:
recommendations['primary'].extend([
'Elbow Method',
'Calinski-Harabasz Index'
])
# Small datasets (n < 1,000)
else:
recommendations['primary'].extend([
'Silhouette Analysis',
'Gap Statistic',
'Dendrogram',
'BIC/AIC'
])
# High-dimensional data (d > 20)
if n_features > 20:
recommendations['avoid'].extend([
'DBSCAN',
'Dendrogram'
])
recommendations['secondary'].append('PCA + any method')
# Non-spherical clusters expected
if cluster_shape == 'arbitrary':
recommendations['primary'].append('DBSCAN')
recommendations['primary'].append('Dendrogram')
recommendations['avoid'].extend([
'Davies-Bouldin Index',
'Calinski-Harabasz Index'
])
# Spherical clusters expected
elif cluster_shape == 'spherical':
recommendations['primary'].extend([
'Calinski-Harabasz Index',
'Davies-Bouldin Index',
'BIC/AIC'
])
# Outliers present
if has_outliers == 'yes':
recommendations['primary'].append('DBSCAN')
recommendations['secondary'].append('Remove outliers first')
# Remove duplicates
recommendations['primary'] = list(set(recommendations['primary']))
recommendations['secondary'] = list(set(recommendations['secondary']))
recommendations['avoid'] = list(set(recommendations['avoid']))
return recommendations
# Example usage
print("Recommendation for large dataset:")
rec1 = recommend_methods(n_samples=50000, n_features=10,
cluster_shape='spherical', time_budget='low')
print("Primary:", rec1['primary'])
print("Avoid:", rec1['avoid'])
print("\nRecommendation for small complex dataset:")
rec2 = recommend_methods(n_samples=500, n_features=5,
cluster_shape='arbitrary', has_outliers='yes')
print("Primary:", rec2['primary'])
Step 3: Workflow Flowchart
START
│
├─ Is n > 10,000?
│ ├─ YES → Use: Elbow, Calinski-Harabasz, Davies-Bouldin
│ └─ NO → Continue
│
├─ Know cluster shape?
│ ├─ Spherical → Calinski-Harabasz, Davies-Bouldin, BIC/AIC
│ ├─ Arbitrary → DBSCAN, Dendrogram, Silhouette
│ └─ Unknown → Try multiple approaches
│
├─ Have outliers?
│ ├─ YES → DBSCAN (auto noise detection)
│ └─ NO → Continue
│
├─ Need statistical rigor?
│ ├─ YES → Gap Statistic, BIC/AIC
│ └─ NO → Elbow, Silhouette
│
├─ Time budget?
│ ├─ Low → Elbow, Calinski-Harabasz
│ ├─ Medium → + Silhouette, BIC/AIC
│ └─ High → + Gap Statistic, Dendrogram
│
└─ ALWAYS: Use 3-5 methods for consensus validation
Production Workflow: From Exploration to Deployment
Here’s a complete, production-ready workflow:
Phase 1: Exploratory Analysis (Quick)
class ClusterExplorer:
"""Quick clustering exploration with multiple methods"""
def __init__(self, X, k_range=range(2, 11)):
self.X = X
self.k_range = k_range
self.results = {}
def quick_screen(self):
"""Fast screening with 3 methods"""
from sklearn.cluster import KMeans
from sklearn.metrics import (calinski_harabasz_score,
davies_bouldin_score)
from kneed import KneeLocator
print("Running quick screening...")
inertias = []
ch_scores = []
db_scores = []
for k in self.k_range:
kmeans = KMeans(n_clusters=k, random_state=42, n_init=10)
labels = kmeans.fit_predict(self.X)
inertias.append(kmeans.inertia_)
ch_scores.append(calinski_harabasz_score(self.X, labels))
db_scores.append(davies_bouldin_score(self.X, labels))
# Find optimal k for each method
kl = KneeLocator(list(self.k_range), inertias,
curve='convex', direction='decreasing')
self.results['elbow'] = kl.elbow
self.results['calinski_harabasz'] = self.k_range[np.argmax(ch_scores)]
self.results['davies_bouldin'] = self.k_range[np.argmin(db_scores)]
print(f" Elbow: k={self.results['elbow']}")
print(f" Calinski-Harabasz: k={self.results['calinski_harabasz']}")
print(f" Davies-Bouldin: k={self.results['davies_bouldin']}")
return self.results
def get_consensus(self):
"""Get consensus k from quick screening"""
from scipy import stats
ks = [k for k in self.results.values() if k is not None]
mode_k = stats.mode(ks, keepdims=True)[0][0]
agreement = ks.count(mode_k) / len(ks)
return mode_k, agreement
# Example usage
explorer = ClusterExplorer(X_scaled)
quick_results = explorer.quick_screen()
consensus_k, agreement = explorer.get_consensus()
print(f"\nQuick consensus: k={consensus_k} ({agreement*100:.0f}% agreement)")
Phase 2: Detailed Validation (If Needed)
class ClusterValidator:
"""Detailed validation with statistical rigor"""
def __init__(self, X, candidate_ks):
self.X = X
self.candidate_ks = candidate_ks
self.results = {}
def detailed_validation(self):
"""Run computationally expensive methods"""
from sklearn.metrics import silhouette_score
from sklearn.mixture import GaussianMixture
print("\nRunning detailed validation...")
for k in self.candidate_ks:
print(f"\nEvaluating k={k}:")
# Silhouette
kmeans = KMeans(n_clusters=k, random_state=42, n_init=10)
labels = kmeans.fit_predict(self.X)
sil = silhouette_score(self.X, labels)
print(f" Silhouette: {sil:.4f}")
# BIC/AIC
gmm = GaussianMixture(n_components=k, random_state=42)
gmm.fit(self.X)
print(f" BIC: {gmm.bic(self.X):.2f}")
print(f" AIC: {gmm.aic(self.X):.2f}")
self.results[k] = {
'silhouette': sil,
'bic': gmm.bic(self.X),
'aic': gmm.aic(self.X)
}
return self.results
def recommend(self):
"""Make final recommendation"""
# Find best k for each metric
best_sil = max(self.results.items(),
key=lambda x: x[1]['silhouette'])[0]
best_bic = min(self.results.items(),
key=lambda x: x[1]['bic'])[0]
best_aic = min(self.results.items(),
key=lambda x: x[1]['aic'])[0]
print("\n" + "="*50)
print("FINAL RECOMMENDATION")
print("="*50)
print(f"Best by Silhouette: k={best_sil}")
print(f"Best by BIC: k={best_bic}")
print(f"Best by AIC: k={best_aic}")
# Consensus
from scipy import stats
ks = [best_sil, best_bic, best_aic]
mode_k = stats.mode(ks, keepdims=True)[0][0]
print(f"\nConsensus recommendation: k={mode_k}")
print("="*50)
return mode_k
# Example usage
if agreement < 0.7: # If quick screening didn't reach consensus
print("\nLow consensus detected. Running detailed validation...")
validator = ClusterValidator(X_scaled,
candidate_ks=[consensus_k-1, consensus_k, consensus_k+1])
detailed_results = validator.detailed_validation()
final_k = validator.recommend()
else:
final_k = consensus_k
print(f"\nStrong consensus from quick screening. Recommended k={final_k}")
Phase 3: Production Deployment
class ProductionClustering:
"""Production-ready clustering with validation"""
def __init__(self, n_clusters, method='kmeans'):
self.n_clusters = n_clusters
self.method = method
self.model = None
self.scaler = StandardScaler()
def fit(self, X):
"""Fit clustering model"""
# Scale features
X_scaled = self.scaler.fit_transform(X)
# Train model
if self.method == 'kmeans':
from sklearn.cluster import KMeans
self.model = KMeans(n_clusters=self.n_clusters,
random_state=42, n_init=10)
elif self.method == 'gmm':
from sklearn.mixture import GaussianMixture
self.model = GaussianMixture(n_components=self.n_clusters,
random_state=42)
elif self.method == 'dbscan':
from sklearn.cluster import DBSCAN
# Parameters should be tuned beforehand
self.model = DBSCAN(eps=0.5, min_samples=4)
self.model.fit(X_scaled)
# Compute quality metrics
labels = self.predict(X)
self.quality_metrics = self._compute_metrics(X_scaled, labels)
return self
def predict(self, X):
"""Predict cluster labels"""
X_scaled = self.scaler.transform(X)
return self.model.fit_predict(X_scaled)
def _compute_metrics(self, X_scaled, labels):
"""Compute clustering quality metrics"""
from sklearn.metrics import (silhouette_score,
davies_bouldin_score,
calinski_harabasz_score)
metrics = {
'silhouette': silhouette_score(X_scaled, labels),
'davies_bouldin': davies_bouldin_score(X_scaled, labels),
'calinski_harabasz': calinski_harabasz_score(X_scaled, labels),
'n_clusters': len(set(labels)) - (1 if -1 in labels else 0)
}
return metrics
def summary(self):
"""Print model summary"""
print("\n" + "="*50)
print("PRODUCTION MODEL SUMMARY")
print("="*50)
print(f"Method: {self.method}")
print(f"Number of clusters: {self.n_clusters}")
print("\nQuality Metrics:")
for metric, value in self.quality_metrics.items():
print(f" {metric}: {value:.4f}")
print("="*50)
# Deploy to production
prod_model = ProductionClustering(n_clusters=final_k, method='kmeans')
prod_model.fit(X)
prod_model.summary()
# Save model
import joblib
joblib.dump(prod_model, 'clustering_model.pkl')
print("\nModel saved to: clustering_model.pkl")
Case Studies: Different Data Types
Let’s see how to apply our framework to different real-world scenarios:
Case Study 1: Customer Segmentation (Tabular Data)
# Simulated customer data
np.random.seed(42)
n_customers = 1000
# Features: age, income, spending_score, tenure
customers = np.column_stack([
np.random.normal(40, 15, n_customers), # age
np.random.normal(60000, 20000, n_customers), # income
np.random.normal(50, 25, n_customers), # spending score
np.random.normal(3, 2, n_customers) # tenure (years)
])
customers_scaled = StandardScaler().fit_transform(customers)
print("Customer Segmentation Analysis")
print("="*50)
# Quick screening
explorer = ClusterExplorer(customers_scaled, k_range=range(2, 8))
results = explorer.quick_screen()
k, agreement = explorer.get_consensus()
print(f"\nRecommended segments: {k}")
print(f"Use case: Create {k} customer personas for targeted marketing")
Case Study 2: Image Compression (High-Dimensional)
# Simulated image pixels (e.g., for color quantization)
np.random.seed(42)
n_pixels = 10000
n_colors = 3 # RGB
# Generate image with 5 dominant colors
image_pixels = np.vstack([
np.random.normal([200, 50, 50], 20, (2000, n_colors)), # Red
np.random.normal([50, 200, 50], 20, (2000, n_colors)), # Green
np.random.normal([50, 50, 200], 20, (2000, n_colors)), # Blue
np.random.normal([200, 200, 50], 20, (2000, n_colors)), # Yellow
np.random.normal([200, 50, 200], 20, (2000, n_colors)) # Magenta
])
print("\nImage Color Quantization")
print("="*50)
# For image compression, use fast methods (millions of pixels)
from sklearn.cluster import MiniBatchKMeans
inertias = []
for k in range(2, 16):
kmeans = MiniBatchKMeans(n_clusters=k, random_state=42, batch_size=1000)
kmeans.fit(image_pixels)
inertias.append(kmeans.inertia_)
kl = KneeLocator(range(2, 16), inertias, curve='convex', direction='decreasing')
print(f"Optimal colors for compression: {kl.elbow}")
print(f"Use case: Reduce image to {kl.elbow} colors for faster transmission")
Case Study 3: Anomaly Detection (With Outliers)
# Simulated network traffic data with anomalies
np.random.seed(42)
n_normal = 900
n_anomaly = 100
# Normal traffic: 2 patterns
normal_traffic = np.vstack([
np.random.normal([100, 50], 10, (450, 2)), # Pattern 1
np.random.normal([200, 150], 15, (450, 2)) # Pattern 2
])
# Anomalous traffic: scattered
anomaly_traffic = np.random.uniform([0, 0], [300, 300], (n_anomaly, 2))
traffic_data = np.vstack([normal_traffic, anomaly_traffic])
traffic_scaled = StandardScaler().fit_transform(traffic_data)
print("\nNetwork Anomaly Detection")
print("="*50)
# DBSCAN excels with outliers
from sklearn.neighbors import NearestNeighbors
k = 5
nbrs = NearestNeighbors(n_neighbors=k).fit(traffic_scaled)
distances, _ = nbrs.kneighbors(traffic_scaled)
distances = np.sort(distances[:, k-1])
# Find elbow in k-distance plot
kl = KneeLocator(range(len(distances)), distances,
curve='convex', direction='increasing')
optimal_eps = distances[kl.elbow] if kl.elbow else 0.3
dbscan = DBSCAN(eps=optimal_eps, min_samples=k)
labels = dbscan.fit_predict(traffic_scaled)
n_clusters = len(set(labels)) - (1 if -1 in labels else 0)
n_noise = list(labels).count(-1)
print(f"Normal traffic patterns detected: {n_clusters}")
print(f"Anomalies detected: {n_noise}")
print(f"Use case: Flag {n_noise} suspicious network connections for review")
Key Takeaways & Best Practices
After this comprehensive journey through 8 clustering validation methods, here are the essential lessons:
1. No Single Method Rules Them All
Different methods excel in different scenarios:
- Fast exploration → Elbow, Calinski-Harabasz
- Statistical rigor → Gap Statistic, BIC/AIC
- Visual interpretation → Dendrogram, Silhouette plots
- Arbitrary shapes → DBSCAN, Dendrogram
- Outlier handling → DBSCAN
Always use 3-5 methods for robust validation.
2. Consensus > Individual Results
# Bad practice
k = elbow_method(X) # Relying on single method
# Good practice
k_elbow = elbow_method(X)
k_silhouette = silhouette_method(X)
k_gap = gap_statistic(X)
k_calinski = calinski_harabasz_method(X)
# Take consensus
from scipy import stats
k_final = stats.mode([k_elbow, k_silhouette, k_gap, k_calinski])[0][0]
3. Domain Knowledge is Crucial
Statistical metrics guide, domain knowledge decides:
Example: Customer segmentation
├─ Methods suggest: k=7
├─ Business constraint: Only 3 marketing teams
└─ Final decision: k=3 (align with resources)
Example: Gene expression clustering
├─ Methods suggest: k=5
├─ Biological knowledge: 3 known cell types
└─ Investigate: k=5 might reveal 2 new subtypes!
4. Data Preprocessing Matters
# Always scale features
from sklearn.preprocessing import StandardScaler
X_scaled = StandardScaler().fit_transform(X)
# Consider dimensionality reduction for high-d data
from sklearn.decomposition import PCA
if X.shape[1] > 20:
pca = PCA(n_components=0.95) # Keep 95% variance
X_reduced = pca.fit_transform(X_scaled)
5. Validate with External Metrics
If you have ground truth labels (even for a subset):
from sklearn.metrics import (adjusted_rand_score,
adjusted_mutual_info_score,
normalized_mutual_info_score)
# Compare predicted vs true labels
ari = adjusted_rand_score(y_true, y_pred)
ami = adjusted_mutual_info_score(y_true, y_pred)
nmi = normalized_mutual_info_score(y_true, y_pred)
print(f"ARI: {ari:.3f}") # 1=perfect, 0=random
print(f"AMI: {ami:.3f}")
print(f"NMI: {nmi:.3f}")
6. Cluster Stability Matters
Test if clusters are stable:
from sklearn.cluster import KMeans
def cluster_stability(X, k, n_runs=50):
"""Test clustering stability across multiple runs"""
labels_list = []
for _ in range(n_runs):
kmeans = KMeans(n_clusters=k, n_init=1) # Single init
labels = kmeans.fit_predict(X)
labels_list.append(labels)
# Average ARI between all pairs
from sklearn.metrics import adjusted_rand_score
scores = []
for i in range(n_runs):
for j in range(i+1, n_runs):
scores.append(adjusted_rand_score(labels_list[i],
labels_list[j]))
avg_stability = np.mean(scores)
print(f"Clustering stability (k={k}): {avg_stability:.3f}")
return avg_stability
# Test k=2 vs k=3
stability_2 = cluster_stability(X_scaled, k=2)
stability_3 = cluster_stability(X_scaled, k=3)
7. Document Your Decision Process
For reproducibility and transparency:
clustering_report = {
'date': '2024-01-29',
'dataset': 'iris',
'n_samples': X.shape[0],
'n_features': X.shape[1],
'methods_used': ['elbow', 'silhouette', 'gap_statistic', 'dbscan'],
'results': {
'elbow': 3,
'silhouette': 2,
'gap_statistic': 3,
'dbscan': 3
},
'consensus': 3,
'final_decision': 3,
'rationale': 'Strong consensus (75%) on k=3. Aligns with domain knowledge.',
'quality_metrics': {
'silhouette': 0.5528,
'davies_bouldin': 0.6619,
'calinski_harabasz': 561.63
}
}
# Save report
import json
with open('clustering_report.json', 'w') as f:
json.dump(clustering_report, f, indent=2)
Final Recommendations: Your Clustering Toolkit
Based on everything we’ve covered, here’s my recommended toolkit for different scenarios:
Scenario 1: Quick Business Analysis
Goal: Get insights fast for presentation
Toolkit:
1. Elbow Method (visualization)
2. Calinski-Harabasz (single number to report)
3. Silhouette plots (show stakeholders cluster quality)
Time: ~5 minutes
Scenario 2: Academic Research
Goal: Publishable, rigorous analysis
Toolkit:
1. Gap Statistic (formal hypothesis test)
2. Silhouette Analysis (detailed validation)
3. BIC/AIC (model selection framework)
4. Stability analysis (show robustness)
Time: ~30 minutes
Include: confidence intervals, sensitivity analysis
Scenario 3: Production ML Pipeline
Goal: Automated, scalable, monitored
Toolkit:
1. Quick screen: Elbow + Calinski-Harabasz
2. If consensus low: Add Gap Statistic
3. Deploy: Production class with monitoring
4. Track: Silhouette score over time (drift detection)
Time: ~10 minutes initial, automated thereafter
Scenario 4: Exploratory Data Analysis
Goal: Understand data structure
Toolkit:
1. Dendrogram (visual hierarchy)
2. DBSCAN (discover arbitrary patterns)
3. Silhouette plots (detailed examination)
4. Multiple k values (explore alternatives)
Time: ~20 minutes
Focus: Understanding, not deciding
Conclusion: The Journey Complete
I hope you found this series of articles is useful and you’ve now mastered 8 different methods for finding the optimal number of clusters, spanning:
- Visual methods: Elbow, Dendrogram
- Geometric methods: Silhouette, Davies-Bouldin
- Statistical methods: Calinski-Harabasz, Gap Statistic
- Probabilistic methods: BIC, AIC
- Density-based methods: DBSCAN
The Meta-Lesson
The ultimate lesson isn’t about any single method - it’s about triangulation. In data science, as in navigation, you get your best bearing by taking readings from multiple angles.
No method is perfect. But together, they’re powerful.
What’s Next for You?
Now that you have this comprehensive toolkit:
- Practice on your own data - Try all 8 methods on a real project
- Build your intuition - Notice which methods agree/disagree and why
- Create your own framework - Adapt recommendations to your domain
- Share your findings - Write about your experiences
Going Deeper
Want to explore further?
Advanced topics to investigate:
- Consensus clustering: Combine multiple clustering results
- Subspace clustering: Find clusters in feature subsets
- Time-series clustering: DTW distance, shape-based methods
- Deep clustering: Neural network-based approaches
- Ensemble methods: Cluster ensembles for robustness
Recommended reading:
- Tibshirani et al. (2001) - Gap Statistic original paper
- Rousseeuw (1987) - Silhouettes original paper
- Ester et al. (1996) - DBSCAN original paper
- “Cluster Analysis” by Everitt et al. - Comprehensive textbook
Series Wrap-Up
This concludes our 3-part series on finding the optimal number of clusters:
- Part 1: Foundation methods - Quick, intuitive validation
- Part 2: Advanced statistics - Rigorous, mathematical approaches
- Part 3: Alternative methods - Flexible, discovery-oriented techniques
Your Feedback Matters
This series was written to bridge the gap between theory and practice. Did it help you? What would you add? What was unclear?
Drop a comment below, or reach out:
- LinkedIn: Randy Frans Fela
- Email: randyrff@gmail.com
Special Thanks
To my colleagues at Jabra who inspired many of these case studies, to the open-source community for incredible tools like scikit-learn, and to you for reading all the way to the end!
Apply This Knowledge
The best way to learn is by doing. Here’s your homework:
- Take a dataset you’re working with
- Apply 4-5 methods from this series
- Document your process
- Share your findings (blog post, talk, or just with your team)
I’d love to see what you discover!
Final Thoughts
In my work at Jabra, evaluating perceptual audio quality metrics, I use clustering daily. The methods in this series aren’t just academic exercises - they’re tools I rely on to make real decisions about product features.
Your journey with clustering is just beginning. These 8 methods are your compass - use them wisely, combine them thoughtfully, and always remember:
“The goal of clustering isn’t to find THE answer - it’s to understand your data well enough to ask better questions.”
Happy clustering! 🎯🚀
Tags: #clustering #machinelearning #datascience #python #unsupervisedlearning #dbscan #hierarchicalclustering #bestpractices #production
Enjoy Reading This Article?
Here are some more articles you might like to read next: